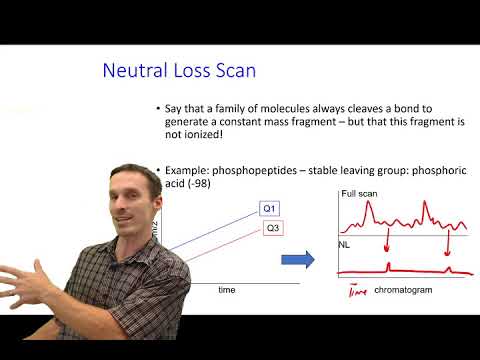
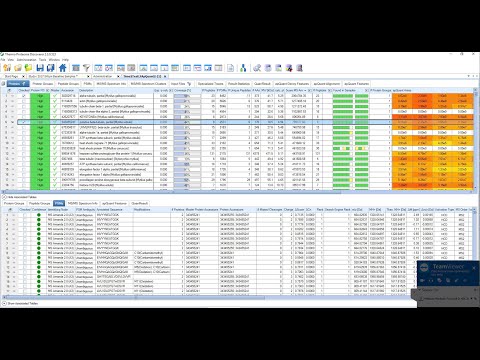
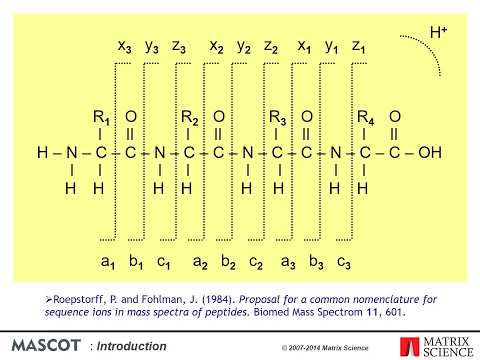
An introduction to computational analysis of mass spectrometry-based proteomics data. In this video, I give a recap of the experimental data before diving into how search databases are created, how spectra are searched against these, how false discovery rates (FDRs) are estimated, and finally the challenges of inferring proteins. If you are not already familiar with MS-based proteomics, I strongly recommend that you watch my short introduction to the core concepts first: 0:00 Introduction: computational proteomics and overview of the presentation. 0:36 Experimental recap: sample preparation, tryptic digest, and MS-based proteomics. 1:05 Search database: sequences, in silico digestion, PTM expansion, and fragment ion prediction 2:20 Spectrum matching: peptide-spectrum matches (PSMs), precursor mass filter, and scoring schemes 3:23 Target-decoy search: decoy spectra, score distributions, and FDR estimation 4:44 Protein inference: equivalent proteins, subset proteins, and protein groups Thank you to David Tabb for his original slide deck that I took inspiration from ( ).
- 6527Просмотров
- 1 год назадОпубликованоLars Juhl Jensen
Protein identification: A deeper dive into analysis of MS-based proteomics data
Похожее видео
Популярное
ЧУПИ В ШКОЛЕ
Две сестры / Дві сестри
малыш вилли
карусель
Universal major 4
СТРАЖИ ПРАВОСУДИЯ 4
Disney channel ukraine
Universal not scary g major 4
6 часть красная гадюка
ЖИЗНЬ ПЕТРОВИЧА В США
красный тарантул часть 3
Universal g major in pp effects
Лупдиду
потерянный снайпер 6 серия
сваты сезон 1 все серии
Aradhana movie
ГРАНЬ ПРАВОСУДИЯ
Обриси
Гранд правосудия 4
Лихач 4сезон 10-12
Красная гадюка часть 4
Valu stupid wife christmas special
Boo boo song Dana
Зворотний напрямок
Две сестры / Дві сестри
малыш вилли
карусель
Universal major 4
СТРАЖИ ПРАВОСУДИЯ 4
Disney channel ukraine
Universal not scary g major 4
6 часть красная гадюка
ЖИЗНЬ ПЕТРОВИЧА В США
красный тарантул часть 3
Universal g major in pp effects
Лупдиду
потерянный снайпер 6 серия
сваты сезон 1 все серии
Aradhana movie
ГРАНЬ ПРАВОСУДИЯ
Обриси
Гранд правосудия 4
Лихач 4сезон 10-12
Красная гадюка часть 4
Valu stupid wife christmas special
Boo boo song Dana
Зворотний напрямок
Новини